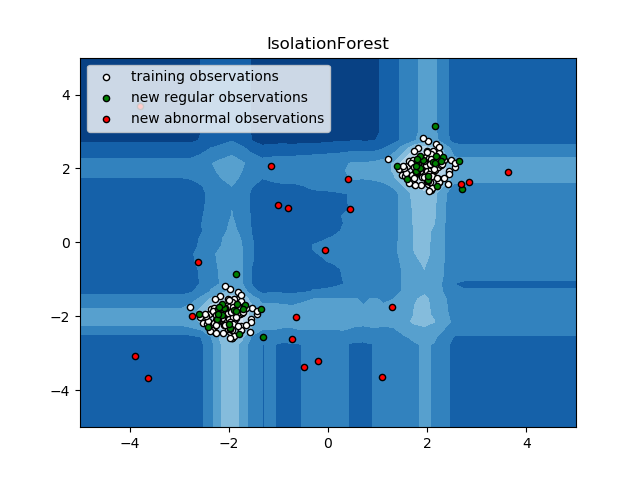
Classification: isolationForest¶
Reference:
scikit learn
https://scikit-learn.org/stable/modules/outlier_detection.html
One efficient way of performing outlier detection in high-dimensional datasets is to use random forests. The ensemble.IsolationForest ‘isolates’ observations by randomly selecting a feature and then randomly selecting a split value between the maximum and minimum values of the selected feature.
Since recursive partitioning can be represented by a tree structure, the number of splittings required to isolate a sample is equivalent to the path length from the root node to the terminating node.
This path length, averaged over a forest of such random trees, is a measure of normality and our decision function.
Random partitioning produces noticeably shorter paths for anomalies. Hence, when a forest of random trees collectively produce shorter path lengths for particular samples, they are highly likely to be anomalies.
The implementation of ensemble.IsolationForest is based on an ensemble of tree.ExtraTreeRegressor. Following Isolation Forest original paper, the maximum depth of each tree is set to where is the number of samples used to build the tree (see (Liu et al., 2008) for more details).
This algorithm is illustrated below.