Kernel Author:
Bhishan Poudel, Ph.D Astrophysics .
Bhishan Poudel, Ph.D Astrophysics .
Data Description¶
In this project, we will predict the probability that an auto insurance policy holder files a claim. This a binary classification problem.
We have more than half a million records and 59 features (including already calculated features).
binary features: _bin
categorical features: _cat
continuous or ordinal feafures: ind, reg, car, calc
missing values: -1
Fullforms
ind = individual
reg = registration
car = car
calc = calculatedThe target columns signifies whether or not a claim was filed for that policy holder.
Evaluation Metric¶
- https://www.kaggle.com/c/porto-seguro-safe-driver-prediction/overview/evaluation
- https://en.wikipedia.org/wiki/Gini_coefficient
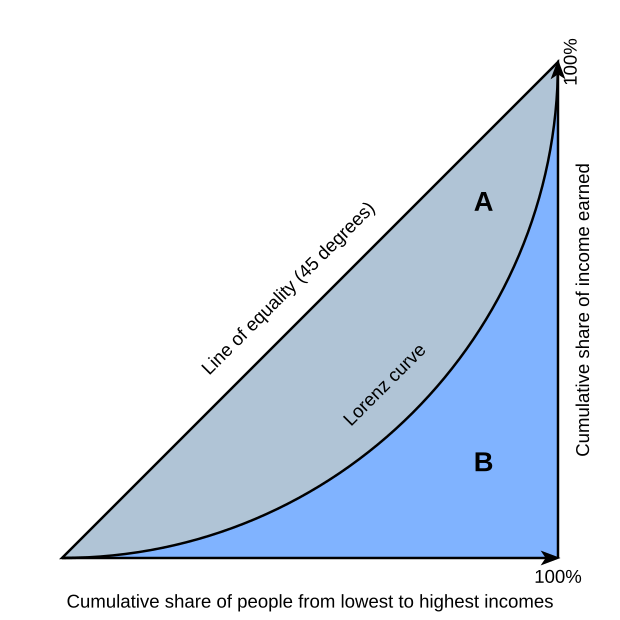
From this graph of wikipedia G = A / (A+B). Gini index varies between 0 and 1. Here we have only binary options: rich and poor.
x-axis= number of people (cumulative sum)
y-axis = total income (cumulative sum)
0 = complete equality of richness
1 = complete inequality of richness
This competition
0 = random guessing
1 = maximum score (also remember 2*1-1 = 1 when maximum auc is 1).If we calculate gini from gini = 2*auc -1 it has range (-1,1).
For AUC:
worst binary classifier AUC = 0.5
perfect binary classifier AUC = 1
If AUC is less than below, simply simply invert 0 <==> 1 then we will get roc auc score between 0.5 and 1.0Imports¶
In [ ]:
import os
import time
import gc
import numpy as np
import pandas as pd
import scipy
from scipy import stats
import seaborn as sns
sns.set(color_codes=True)
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
time_start_notebook = time.time()
SEED=100
print([(x.__name__,x.__version__) for x in [np, pd,sns,matplotlib]])
from scipy import sparse as ssp
from sklearn.model_selection import StratifiedKFold
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
In [ ]:
# Google colab
In [ ]:
%%capture
# capture will not print in notebook
import os
import sys
ENV_COLAB = 'google.colab' in sys.modules
if ENV_COLAB:
#### print
print('Environment: Google Colaboratory.')
# NOTE: If we update modules in gcolab, we need to restart runtime.
# When using colab for xgboost or deep learning, we can use gpu.
Useful Functions¶
In [ ]:
df_eval = pd.DataFrame({'Model': [],
'Description':[],
'Accuracy':[],
'Precision':[],
'Recall':[],
'F1':[],
'AUC':[],
'NormlaizedGini': []
})
Evaluation Metric¶
In [ ]:
# @numba.jit fails and falls back to object mode.
def eval_gini(y_true, y_prob):
y_true = np.asarray(y_true)
y_true = y_true[np.argsort(y_prob)]
ntrue = 0
gini = 0
delta = 0
n = len(y_true)
for i in range(n-1, -1, -1):
y_i = y_true[i]
ntrue += y_i
gini += y_i * delta
delta += 1 - y_i
gini = 1 - 2 * gini / (ntrue * (n - ntrue))
return gini
def gini_xgb(preds, dtrain):
labels = dtrain.get_label()
gini_score = -eval_gini(labels, preds)
return [('gini', gini_score)]
Load the data¶
In [ ]:
df = pd.read_csv('https://github.com/bhishanpdl/Datasets/blob/master/'
'Porto_seguro_safe_driver_prediction/train.csv.zip?raw=true',
compression='zip',na_values=None)
print(df.shape)
df.head()
Out[ ]:
In [ ]:
target = 'target'
Data Processing¶
In [ ]:
# all features except target
cols_all= df.columns.drop(target).to_list()
# categorical features except later created count
cols_cat = [c for c in cols_all if ('cat' in c and 'count' not in c)]
# we exclude calc features in numeric features
cols_num = [c for c in cols_all if ('cat' not in c and 'calc' not in c)]
print(cols_num)
In [ ]:
# missing count
df['missing'] = df.eq(-1).sum(axis=1).astype(float)
cols_num.append('missing')
# individual features
cols_ind = [c for c in cols_all if 'ind' in c]
df['ind_concat'] = df[cols_ind].astype(str).agg('_'.join,axis=1)
# cat count features from whole data
cols_cat_count = []
for col in cols_cat + ['ind_concat']:
d = df[col].value_counts().to_dict()
df[f'{col}_count'] = df[col].apply(lambda x:d.get(x,0))
cols_cat_count.append(f'{col}_count')
# after creating count of ind concat, drop it
df = df.drop('ind_concat',axis=1)
# combination features
combs = [
('ps_reg_01', 'ps_car_02_cat'),
('ps_reg_01', 'ps_car_04_cat'),
]
cols_comb = []
for (f1, f2) in combs:
le = LabelEncoder()
name = f1 + "_plus_" + f2
df[name] = df[f1].astype(str) + "_" + df[f2].astype(str)
le.fit(df[name].tolist()) # use whole data column
df[name] = le.transform(df[name].tolist()) # replace column name
cols_comb.append(name)
# one hot encoding
# df = pd.get_dummies(df, columns=cols_cat, drop_first=True)
# Here, we will do target encoding of categorical variables instead of OHE.
# we will also keep all these categorical features as well as encoded targets.
Train-test Split with Stratify¶
In [ ]:
from sklearn.model_selection import train_test_split
# note: when using cross validation later, we dont need train-valid split here.
df_Xtrain, df_Xtest, ser_ytrain, ser_ytest = train_test_split(
df.drop(target,axis=1),df[target],
test_size=0.2,random_state=SEED, stratify=df[target])
# backup and delete id
cols_drop = ['id']
train_id = df_Xtrain[cols_drop]
test_id = df_Xtest[cols_drop]
df_Xtrain = df_Xtrain.drop(cols_drop,axis=1)
df_Xtest = df_Xtest.drop(cols_drop,axis=1)
Xtrain = df_Xtrain.to_numpy()
ytrain = ser_ytrain.to_numpy().ravel()
Xtest = df_Xtest.to_numpy()
ytest = ser_ytest.to_numpy().ravel()
# make sure no nans and no strings
print(Xtrain.sum().sum())
In [ ]:
ser_ytest.value_counts(normalize=True)
Out[ ]:
Target Encoding¶
In [ ]:
def add_noise(series, noise_level):
return series * (1 + noise_level * np.random.randn(len(series)))
def target_encode(trn_series=None,
val_series=None,
tst_series=None,
target=None,
min_samples_leaf=1,
smoothing=1,
noise_level=0):
"""
Target encoding for categorical features.
Parameters
-----------
trn_series : pd.Series
One pandas categorical series from training set.
val_series : pd.Series
One pandas categorical series from validation set.
val_series : pd.Series
One pandas categorical series from test set.
target : pd.Series
Target pandas series.
min_samples_leaf: int
Minimum samples to take category average into account
smoothing: int
Smoothing effect to balance categorical average vs prior
noise: float
Introduce noise to avoid data leakage.
Notes
------
Smoothing is computed like in the following paper by Daniele Micci-Barreca
https://kaggle2.blob.core.windows.net/forum-message-attachments/
225952/7441/high%20cardinality%20categoricals.pdf
References
-----------
https://www.kaggle.com/aharless/xgboost-cv-lb-284
"""
assert len(trn_series) == len(target)
assert trn_series.name == tst_series.name
temp = pd.concat([trn_series, target], axis=1)
# Compute target mean
averages = temp.groupby(by=trn_series.name)[target.name].agg(["mean", "count"])
# Compute smoothing
smoothing = 1 / (1 + np.exp(-(averages["count"] - min_samples_leaf) / smoothing))
# Apply average function to all target data
prior = target.mean()
# The bigger the count the less full_avg is taken into account
averages[target.name] = prior * (1 - smoothing) + averages["mean"] * smoothing
averages.drop(["mean", "count"], axis=1, inplace=True)
# Apply averages to trn and tst series
ft_trn_series = pd.merge(
trn_series.to_frame(trn_series.name),
averages.reset_index().rename(columns={'index': target.name,
target.name: 'average'}),
on=trn_series.name,
how='left')['average'].rename(trn_series.name + '_mean').fillna(prior)
# pd.merge does not keep the index so restore it
ft_trn_series.index = trn_series.index
ft_val_series = pd.merge(
val_series.to_frame(val_series.name),
averages.reset_index().rename(columns={'index': target.name,
target.name: 'average'}),
on=val_series.name,
how='left')['average'].rename(trn_series.name + '_mean').fillna(prior)
# pd.merge does not keep the index so restore it
ft_val_series.index = val_series.index
ft_tst_series = pd.merge(
tst_series.to_frame(tst_series.name),
averages.reset_index().rename(columns={'index': target.name,
target.name: 'average'}),
on=tst_series.name,
how='left')['average'].rename(trn_series.name + '_mean').fillna(prior)
# pd.merge does not keep the index so restore it
ft_tst_series.index = tst_series.index
return [add_noise(ft_trn_series, noise_level),
add_noise(ft_val_series, noise_level),
add_noise(ft_tst_series, noise_level)]
Modelling: XGBoost¶
In [ ]:
import joblib
import xgboost as xgb
from sklearn.model_selection import StratifiedKFold
from lightgbm import LGBMClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score,precision_score, recall_score
from sklearn.metrics import f1_score, roc_auc_score
In [ ]:
# xgb.XGBClassifier?
# https://xgboost.readthedocs.io/en/latest/parameter.html
In [ ]:
TREE_METHOD = "gpu_hist" if ENV_COLAB else 'auto'
print(TREE_METHOD)
In [ ]:
model = xgb.XGBClassifier(n_jobs=-1, random_state=SEED,
tree_method = TREE_METHOD,
objective='binary:logistic')
model
Out[ ]:
In [ ]:
# fit the model
model.fit(df_Xtrain, ser_ytrain)
# predictions
skf = StratifiedKFold(n_splits=2,shuffle=True,random_state=SEED)
ypreds_cv = cross_val_predict(model, df_Xtest, ser_ytest, cv=skf)
ypreds = ypreds_cv
# model evaluation
average = 'binary'
row_eval = ['Xgboost','default, no target encoding',
accuracy_score(ytest, ypreds),
precision_score(ytest, ypreds, average=average),
recall_score(ytest, ypreds, average=average),
f1_score(ytest, ypreds, average=average),
roc_auc_score(ytest, ypreds),
2 * roc_auc_score(ytest, ypreds) -1,
]
df_eval.loc[len(df_eval)] = row_eval
df_eval = df_eval.drop_duplicates()
display(df_eval)
SKF-CV and Target Encoding¶
In [ ]:
time_start = time.time()
K = 5
skf = StratifiedKFold(n_splits=K, random_state=SEED, shuffle=True)
MAX_ROUNDS = 10
EARLY_STOPPING_ROUNDS = 5
# MAX_ROUNDS = 400
# EARLY_STOPPING_ROUNDS = 50
LEARNING_RATE = 0.07
OPTIMIZE_ROUNDS = False # model fit on eval set and use early stopping
METRIC_NAME = 'Gini'
EVAL_METRIC = gini_xgb
# METRIC_NAME = 'auc'
# EVAL_METRIC = 'auc' # note: normalizedGini = 2*auc -1
# model.best_score will give auc if eval metric is auc.
ser_vdprobs = ser_ytrain * 0
ser_vdpreds = ser_ytrain * 0 # all index of validation makes same as train
txprobs = ser_ytest.to_numpy().ravel() * 0.0 # make it zero, to add values from CV
model = xgb.XGBClassifier(
tree_method=TREE_METHOD,
n_estimators=MAX_ROUNDS,
max_depth=4,
objective="binary:logistic",
learning_rate=LEARNING_RATE,
subsample=.8,
min_child_weight=6,
colsample_bytree=.8,
scale_pos_weight=1.6,
gamma=10,
reg_alpha=8,
n_jobs=-1,
reg_lambda=1.3,
)
best_thr_lst = []
for i, (idx_tr, idx_vd) in enumerate(skf.split(Xtrain, ytrain)):
# print
print( "\nFold ", i)
# data for this fold
df_Xtr = df_Xtrain.iloc[idx_tr,:].copy()
ser_ytr = ser_ytrain.iloc[idx_tr].copy()
df_Xvd = df_Xtrain.iloc[idx_vd,:].copy()
ser_yvd = ser_ytrain.iloc[idx_vd].copy()
df_Xtx = df_Xtest.copy() # we add target encoding features to test
# target encode (do inside validation, not outside, to avoid data leakage.)
for col in cols_cat:
(df_Xtr[col + "_avg"], df_Xvd[col + "_avg"], df_Xtx[col + "_avg"]) = \
target_encode(trn_series=df_Xtr[col],
val_series=df_Xvd[col],
tst_series=df_Xtx[col],
target=ser_ytr,
min_samples_leaf=200,
smoothing=10,
noise_level=0
)
# fit model for this fold
if OPTIMIZE_ROUNDS:
eval_set = [(df_Xvd,ser_yvd)]
fit_model = model.fit( df_Xtr, ser_ytr,
eval_set=eval_set,
eval_metric=EVAL_METRIC,
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
verbose=False
)
print(" Best N trees : ", model.best_ntree_limit )
print(f" Best {METRIC_NAME} : ", model.best_score )
print()
else:
fit_model = model.fit(df_Xtr, ser_ytr )
# valid probs for this fold
vdprobs = fit_model.predict_proba(df_Xvd)[:,1]
print( " Gini (from probs) : ", eval_gini(ser_yvd, vdprobs) )
# find the best threshold using validation data
thresholds = np.arange(0, 1, 0.001)
# using eval_gini is too slow, alternatively we can use auc
# and assume that both have the same best threshold
# note that: gini = 2*auc - 1
#
# gini_scores = [eval_gini(ser_yvd, [0 if i <= thr else 1 for i in vdprobs])
# for thr in thresholds]
# idx = np.argmax(gini_scores)
# best_thr = thresholds[idx]
# best_gini = gini_scores[idx]
# using auc instead of gini to find best threshold
auc_scores = [roc_auc_score(ser_yvd, [0 if i <= thr else 1 for i in vdprobs])
for thr in thresholds]
idx = np.argmax(auc_scores)
best_thr = thresholds[idx]
best_auc = auc_scores[idx]
best_thr_lst.append(best_thr)
best_auc = auc_scores[idx]
vdpreds = [0 if i <= best_thr else 1 for i in vdprobs]
print(f' Best threshold : {best_thr:.3f}')
print(f' Best AUC : {best_auc:.5f}')
print( " Gini (from preds) : ", eval_gini(ser_yvd, vdpreds) )
ser_vdprobs.iloc[idx_vd] = vdprobs
ser_vdpreds.iloc[idx_vd] = vdpreds
# accumulate probs
txprobs += fit_model.predict_proba(df_Xtx)[:,1] # test probs
# clean memory
del df_Xtr, ser_ytr, df_Xvd, ser_yvd, df_Xtx
# time taken
time_taken = time.time() - time_start
h,m = divmod(time_taken,60*60)
print(' Time taken : {:.0f} hr '\
'{:.0f} min {:.0f} secs'.format(h, *divmod(m,60)))
txprobs /= K # avg test probs
best_thr = np.mean(best_thr_lst)
txpreds = [0 if i <= best_thr else 1 for i in txprobs]
print()
print(f'Best thresholds for fold : {best_thr_lst}')
print("Gini for full training set (from probs) : ",
eval_gini(ser_ytrain, ser_vdprobs))
print("Gini for full training set (from preds): ",
eval_gini(ser_ytrain, ser_vdpreds))
# predictions
ypreds = txpreds
# model evaluation
average = 'binary'
row_eval = ['Xgboost','skf cv, cats + target encoding',
accuracy_score(ytest, ypreds),
precision_score(ytest, ypreds, average=average),
recall_score(ytest, ypreds, average=average),
f1_score(ytest, ypreds, average=average),
roc_auc_score(ytest, ypreds),
2 * roc_auc_score(ytest, ypreds) -1,
]
df_eval.loc[len(df_eval)] = row_eval
df_eval = df_eval.drop_duplicates()
display(df_eval)
Selected Features¶
In [ ]:
cols_select = ["ps_car_13","ps_reg_03","ps_ind_05_cat","ps_ind_03","ps_ind_15",
"ps_reg_02","ps_car_14","ps_car_12","ps_car_01_cat","ps_car_07_cat",
"ps_ind_17_bin","ps_car_03_cat","ps_reg_01","ps_car_15","ps_ind_01",
"ps_ind_16_bin","ps_ind_07_bin","ps_car_06_cat","ps_car_04_cat","ps_ind_06_bin",
"ps_car_09_cat","ps_car_02_cat","ps_ind_02_cat","ps_car_11","ps_car_05_cat",
"ps_calc_09","ps_calc_05","ps_ind_08_bin","ps_car_08_cat","ps_ind_09_bin",
"ps_ind_04_cat","ps_ind_18_bin","ps_ind_12_bin","ps_ind_14"]
In [ ]:
cols_select = cols_select + cols_comb
In [ ]:
df_Xtrain = df_Xtrain[cols_select]
df_Xtest = df_Xtest[cols_select]
Xtrain = df_Xtrain.to_numpy()
ytrain = ser_ytrain.to_numpy().ravel()
Xtest = df_Xtest.to_numpy()
ytest = ser_ytest.to_numpy().ravel()
# make sure no nans and no strings
print(Xtrain.sum().sum())
In [ ]:
cols_cat = [i for i in df_Xtrain.columns if i.endswith('_cat')]
In [ ]:
time_start = time.time()
K = 5
skf = StratifiedKFold(n_splits=K, random_state=SEED, shuffle=True)
MAX_ROUNDS = 10
EARLY_STOPPING_ROUNDS = 5
# MAX_ROUNDS = 400
# EARLY_STOPPING_ROUNDS = 50
LEARNING_RATE = 0.07
OPTIMIZE_ROUNDS = False # model fit on eval set and use early stopping
METRIC_NAME = 'Gini'
EVAL_METRIC = gini_xgb
# METRIC_NAME = 'auc'
# EVAL_METRIC = 'auc' # note: normalizedGini = 2*auc -1
# model.best_score will give auc if eval metric is auc.
ser_vdprobs = ser_ytrain * 0
ser_vdpreds = ser_ytrain * 0 # all index of validation makes same as train
txprobs = ser_ytest.to_numpy().ravel() * 0.0 # make it zero, to add values from CV
model = xgb.XGBClassifier(tree_method=TREE_METHOD,
n_estimators=MAX_ROUNDS,max_depth=4,objective="binary:logistic",
learning_rate=LEARNING_RATE, subsample=.8,min_child_weight=6,
colsample_bytree=.8,scale_pos_weight=1.6,gamma=10,reg_alpha=8,
n_jobs=-1,reg_lambda=1.3)
best_thr_lst = []
for i, (idx_tr, idx_vd) in enumerate(skf.split(Xtrain, ytrain)):
# print
print( "\nFold ", i)
# data for this fold
df_Xtr = df_Xtrain.iloc[idx_tr,:].copy()
ser_ytr = ser_ytrain.iloc[idx_tr].copy()
df_Xvd = df_Xtrain.iloc[idx_vd,:].copy()
ser_yvd = ser_ytrain.iloc[idx_vd].copy()
df_Xtx = df_Xtest.copy() # we add target encoding features to test
# target encode (do inside validation, not outside, to avoid data leakage.)
for col in cols_cat:
(df_Xtr[col + "_avg"], df_Xvd[col + "_avg"], df_Xtx[col + "_avg"]) = \
target_encode(trn_series=df_Xtr[col],
val_series=df_Xvd[col],
tst_series=df_Xtx[col],
target=ser_ytr,
min_samples_leaf=200,
smoothing=10,
noise_level=0
)
# fit model for this fold
if OPTIMIZE_ROUNDS:
eval_set = [(df_Xvd,ser_yvd)]
fit_model = model.fit( df_Xtr, ser_ytr,
eval_set=eval_set,
eval_metric=EVAL_METRIC,
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
verbose=False
)
print(" Best N trees : ", model.best_ntree_limit )
print(f" Best {METRIC_NAME} : ", model.best_score )
print()
else:
fit_model = model.fit(df_Xtr, ser_ytr )
# valid probs for this fold
vdprobs = fit_model.predict_proba(df_Xvd)[:,1]
print( " Gini (from probs) : ", eval_gini(ser_yvd, vdprobs) )
# find the best threshold using validation data
thresholds = np.arange(0, 1, 0.001)
# using eval_gini is too slow, alternatively we can use auc
# and assume that both have the same best threshold
# note that: gini = 2*auc - 1
#
# gini_scores = [eval_gini(ser_yvd, [0 if i <= thr else 1 for i in vdprobs])
# for thr in thresholds]
# idx = np.argmax(gini_scores)
# best_thr = thresholds[idx]
# best_gini = gini_scores[idx]
# using auc instead of gini to find best threshold
auc_scores = [roc_auc_score(ser_yvd, [0 if i <= thr else 1 for i in vdprobs])
for thr in thresholds]
idx = np.argmax(auc_scores)
best_thr = thresholds[idx]
best_auc = auc_scores[idx]
best_thr_lst.append(best_thr)
best_auc = auc_scores[idx]
vdpreds = [0 if i <= best_thr else 1 for i in vdprobs]
print(f' Best threshold : {best_thr:.3f}')
print(f' Best AUC : {best_auc:.5f}')
print( " Gini (from preds) : ", eval_gini(ser_yvd, vdpreds) )
ser_vdprobs.iloc[idx_vd] = vdprobs
ser_vdpreds.iloc[idx_vd] = vdpreds
# accumulate probs
txprobs += fit_model.predict_proba(df_Xtx)[:,1] # test probs
# clean memory
del df_Xtr, ser_ytr, df_Xvd, ser_yvd, df_Xtx
# time taken
time_taken = time.time() - time_start
h,m = divmod(time_taken,60*60)
print(' Time taken : {:.0f} hr '\
'{:.0f} min {:.0f} secs'.format(h, *divmod(m,60)))
txprobs /= K # avg test probs
best_thr = np.mean(best_thr_lst)
txpreds = [0 if i <= best_thr else 1 for i in txprobs]
print()
print(f'Best thresholds for fold : {best_thr_lst}')
print("Gini for full training set (from probs) : ",
eval_gini(ser_ytrain, ser_vdprobs))
print("Gini for full training set (from preds): ",
eval_gini(ser_ytrain, ser_vdpreds))
# predictions
ypreds = txpreds
# model evaluation
average = 'binary'
row_eval = ['Xgboost','skf cv, cats + target encoding, few features',
accuracy_score(ytest, ypreds),
precision_score(ytest, ypreds, average=average),
recall_score(ytest, ypreds, average=average),
f1_score(ytest, ypreds, average=average),
roc_auc_score(ytest, ypreds),
2 * roc_auc_score(ytest, ypreds) -1,
]
df_eval.loc[len(df_eval)] = row_eval
df_eval = df_eval.drop_duplicates()
display(df_eval)