Kernel Author:
Bhishan Poudel, Ph.D Astrophysics .
Bhishan Poudel, Ph.D Astrophysics .
Data Description¶
In this project, we will predict the probability that an auto insurance policy holder files a claim. This a binary classification problem.
We have more than half a million records and 59 features (including already calculated features).
binary features: _bin
categorical features: _cat
continuous or ordinal feafures: ind, reg, car, calc
missing values: -1
Fullforms
ind = individual
reg = registration
car = car
calc = calculatedThe target columns signifies whether or not a claim was filed for that policy holder.
Evaluation Metric¶
- https://www.kaggle.com/c/porto-seguro-safe-driver-prediction/overview/evaluation
- https://en.wikipedia.org/wiki/Gini_coefficient
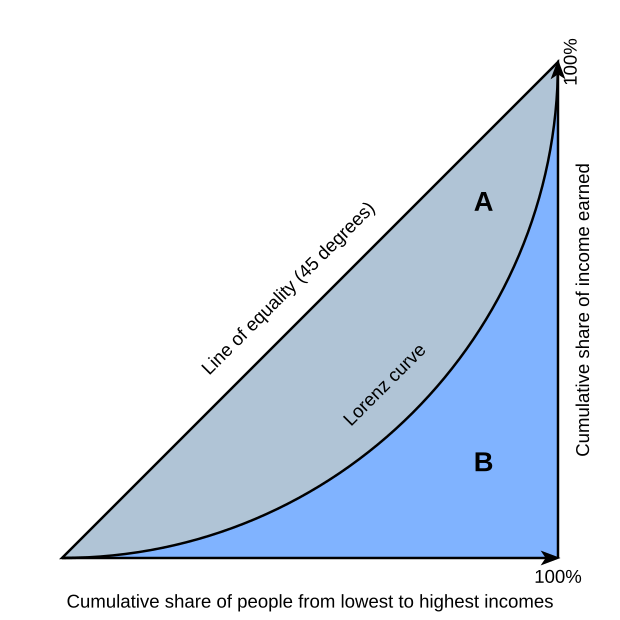
From this graph of wikipedia G = A / (A+B). Gini index varies between 0 and 1. Here we have only binary options: rich and poor.
x-axis= number of people (cumulative sum)
y-axis = total income (cumulative sum)
0 = complete equality of richness
1 = complete inequality of richness
This competition
0 = random guessing
1 = maximum score (also remember 2*1-1 = 1 when maximum auc is 1).If we calculate gini from gini = 2*auc -1 it has range (-1,1).
For AUC:
worst binary classifier AUC = 0.5
perfect binary classifier AUC = 1
If AUC is less than below, simply simply invert 0 <==> 1 then we will get roc auc score between 0.5 and 1.0Imports¶
In [1]:
import os
import time
import gc
import numpy as np
import pandas as pd
import scipy
from scipy import stats
import seaborn as sns
sns.set(color_codes=True)
import matplotlib
import matplotlib.pyplot as plt
from pprint import pprint
%matplotlib inline
time_start_notebook = time.time()
SEED=100
print([(x.__name__,x.__version__) for x in [np, pd,sns,matplotlib]])
from scipy import sparse as ssp
from sklearn.model_selection import StratifiedKFold
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OneHotEncoder
In [2]:
# Google colab
In [26]:
%%capture
# capture will not print in notebook
import os
import sys
ENV_COLAB = 'google.colab' in sys.modules
if ENV_COLAB:
# deep learning
!pip install lrcurve
#### print
print('Environment: Google Colaboratory.')
# NOTE: If we update modules in gcolab, we need to restart runtime.
Useful Functions¶
In [4]:
df_eval = pd.DataFrame({'Model': [],
'Description':[],
'Accuracy':[],
'Precision':[],
'Recall':[],
'F1':[],
'AUC':[],
'NormalizedGini': []
})
Load the data¶
In [5]:
df = pd.read_csv('https://github.com/bhishanpdl/Datasets/blob/master/'
'Porto_seguro_safe_driver_prediction/train.csv.zip?raw=true',compression='zip')
print(df.shape)
# for neural nets, make the data small
# df = df.sample(frac=0.01,random_state=SEED)
df.head()
Out[5]:
In [6]:
"""
Comment about file size:
The data is large, it has 595k records and 59 features.
ps = porto seguro
_bin = binary feature
_cat = categorical feature
continuous or ordinal: ind, reg, car, calc
""";
In [7]:
target = 'target'
Data Processing¶
In [8]:
# all features except target
cols_all= df.columns.drop(target).to_list()
# categorical features except later created count
cols_cat = [c for c in cols_all if ('cat' in c and 'count' not in c)]
# we exclude calc features in numeric features
cols_num = [c for c in cols_all if ('cat' not in c and 'calc' not in c)]
print(cols_num)
Train-test Split with Stratify¶
In [9]:
from sklearn.model_selection import train_test_split
df_Xtrain, df_Xtest, ser_ytrain, ser_ytest = train_test_split(
df.drop(target,axis=1),df[target],
test_size=0.2,random_state=SEED, stratify=df[target])
# backup and delete id
cols_drop = ['id']
train_id = df_Xtrain[cols_drop]
test_id = df_Xtest[cols_drop]
df_Xtrain = df_Xtrain.drop(cols_drop,axis=1)
df_Xtest = df_Xtest.drop(cols_drop,axis=1)
Xtrain = df_Xtrain.to_numpy()
ytrain = ser_ytrain.to_numpy().ravel()
Xtest = df_Xtest.to_numpy()
ytest = ser_ytest.to_numpy().ravel()
# make sure no nans and no strings
print(Xtrain.sum().sum())
Training Data¶
In [10]:
pd.set_option('display.max_columns',250)
df_Xtrain.head()
Out[10]:
In [11]:
# df_Xtrain.columns # make sure there are no id and index
In [12]:
Xtr = Xtrain
Xtx = Xtest
ytr = ytrain
ytx = ytest
print(Xtr.shape, Xtx.shape)
In [13]:
ser_ytest.value_counts(normalize=True)
Out[13]:
Evaluation Metric¶
In [14]:
#gini scoring function from kernel at:
#https://www.kaggle.com/tezdhar/faster-gini-calculation
def ginic(actual, pred):
n = len(actual)
a_s = actual[np.argsort(pred)]
a_c = a_s.cumsum()
giniSum = a_c.sum() / a_c[-1] - (n + 1) / 2.0
return giniSum / n
def gini_normalizedc(a, p):
return ginic(a, p) / ginic(a, a)
Data processing¶
In [15]:
# remove calc features
cols_use = [c for c in df_Xtrain.columns if (not c.startswith('ps_calc_'))]
df_Xtrain = df_Xtrain[cols_use]
df_Xtest = df_Xtest[cols_use]
Build embedding network¶
In [16]:
# if nunique is >2 make embed dict of cats
cols_cat = [i for i in df_Xtrain.columns if i.endswith('_cat')]
# print(cols_cat)
df_emb = df_Xtrain[cols_cat].nunique().loc[lambda x: x>2].to_frame('nunique')
# df_emb
In [17]:
df_emb['nunique'].values
Out[17]:
In [18]:
df_emb['size'] = [ (5,3),(3,2),(8,5),(13,7), (3,2),
(3,2), (10,5), (3,2), (18,8), (3,2),
(6,3), (3,2), (104,10) ]
df_emb
Out[18]:
In [19]:
dict_emb = df_emb['size'].to_dict()
dict_emb
Out[19]:
In [20]:
def build_embedding_network():
"""Build the embedding network.
Parameters
-----------
dict_emb: embedding dict eg. {'mycol': (10,8), 'mycols2': (3,2)}
mycol has originally 8 unique categorical values but
we want to embed 8 dimensional space.
Usage
------
NN = build_embedding_network()
NN.fit(proc_Xtr, ser_ytr.values)
"""
inputs = []
embeddings = []
for key in dict_emb.keys():
input = Input(shape=(1,))
x,y = dict_emb[key]
embedding = Embedding(x, y, input_length=1)(input)
embedding = Reshape(target_shape=(y,))(embedding)
inputs.append(input)
embeddings.append(embedding)
input_numeric = Input(shape=(24,))
embedding_numeric = Dense(16)(input_numeric)
inputs.append(input_numeric)
embeddings.append(embedding_numeric)
x = Concatenate()(embeddings)
x = Dense(80, activation='relu')(x)
x = Dropout(.35)(x)
x = Dense(20, activation='relu')(x)
x = Dropout(.15)(x)
x = Dense(10, activation='relu')(x)
x = Dropout(.15)(x)
output = Dense(1, activation='sigmoid')(x)
model = Model(inputs, output)
model.compile(loss='binary_crossentropy', optimizer='adam')
return model
In [21]:
# converting data to list format to match the network structure
def preproc(df_Xtr, df_Xvd, df_Xtx):
"""Preprocessing data for neural network fitting.
Parameters
-----------
df_Xtr: training dataframe
df_Xvd: validation dataframe
df_Xtx: test dataframe
dict_emb: embedding dict eg. {'mycol': (10,8), 'mycols2': (3,2)}
mycol has originally 8 unique categorical values but
we want to embed 8 dimensional space.
Usage
-----
proc_Xtr, proc_Xvd, proc_Xtx = preproc(df_Xtr,df_Xvd, df_Xtx)
NN = build_embedding_network()
NN.fit(proc_Xtr, ser_ytr.values)
"""
input_list_train = []
input_list_val = []
input_list_test = []
# the cols to be embedded: rescaling to range [0, # values)
for c in dict_emb.keys():
raw_vals = np.unique(df_Xtr[c])
val_map = {}
for i in range(len(raw_vals)):
val_map[raw_vals[i]] = i
input_list_train.append(df_Xtr[c].map(val_map).values)
input_list_val.append(df_Xvd[c].map(val_map).fillna(0).values)
input_list_test.append(df_Xtx[c].map(val_map).fillna(0).values)
# the rest of the columns
other_cols = [c for c in df_Xtr.columns if (not c in dict_emb.keys())]
input_list_train.append(df_Xtr[other_cols].values)
input_list_val.append(df_Xvd[other_cols].values)
input_list_test.append(df_Xtx[other_cols].values)
return input_list_train, input_list_val, input_list_test
Modelling: Keras Entity Embeddings¶
In [28]:
from keras.models import Model
from keras.layers import Input, Dense, Concatenate, Reshape, Dropout
from keras.layers.embeddings import Embedding
from sklearn.model_selection import StratifiedKFold
from lrcurve import KerasLearningCurve
In [30]:
K = 5 # number of folds
runs_per_fold = 3
n_epochs = 15 # make it large eg. 100
cv_ginis = []
trprobs = np.zeros(np.shape(df_Xtrain)[0]) # Ntrain rows (full validation set)
txprobs = np.zeros((np.shape(df_Xtest)[0],K)) # Ntest rows, K columns
skf = StratifiedKFold(n_splits=K, random_state=SEED, shuffle=True)
time_start = time.time()
for i, (idx_tr, idx_vd) in enumerate(skf.split(df_Xtrain.to_numpy(),
ser_ytrain.to_numpy())):
# print
print( "\nFold ", i)
# data for this fold
df_Xtr = df_Xtrain.iloc[idx_tr,:].copy()
ser_ytr = ser_ytrain.iloc[idx_tr].copy()
df_Xvd = df_Xtrain.iloc[idx_vd,:].copy()
ser_yvd = ser_ytrain.iloc[idx_vd].copy()
df_Xtx = df_Xtest.copy()
# upsampling
pos = (ser_ytr == 1)
# add positive examples
df_Xtr = pd.concat([df_Xtr, df_Xtr.loc[pos]], axis=0)
ser_ytr = pd.concat([ser_ytr, ser_ytr.loc[pos]], axis=0)
# shuffle data
idx = np.arange(len(df_Xtr))
np.random.shuffle(idx)
df_Xtr = df_Xtr.iloc[idx]
ser_ytr = ser_ytr.iloc[idx]
# preprocessing
proc_Xtr, proc_Xvd, proc_Xtx = preproc(df_Xtr,df_Xvd, df_Xtx)
# track oof prediction for cv scores
vdprobs = 0 # we must init it
for j in range(runs_per_fold):
NN = build_embedding_network()
NN.fit(proc_Xtr, ser_ytr.values,
validation_data = (proc_Xvd, ser_yvd.to_numpy()),
callbacks=[KerasLearningCurve()],
epochs=n_epochs,batch_size=4096, verbose=0)
vdprobs += NN.predict(proc_Xvd)[:,0] / runs_per_fold
txprobs[:,i] += NN.predict(proc_Xtx)[:,0] / runs_per_fold
trprobs[idx_vd] += vdprobs # train is the full validation set
cv_gini = gini_normalizedc(ser_yvd.values, vdprobs)
cv_ginis.append(cv_gini)
print (f'\n cv gini: {cv_gini:.5f}')
# clean memory
del df_Xtr, df_Xvd, df_Xtx, proc_Xtr, proc_Xvd, proc_Xtx
# time taken
time_taken = time.time() - time_start
h,m = divmod(time_taken,60*60)
print(' Time taken : {:.0f} hr '\
'{:.0f} min {:.0f} secs'.format(h, *divmod(m,60)))
# outside the loop
txprobs = np.mean(txprobs, axis=1)
print('Mean out of fold gini: %.5f' % np.mean(cv_ginis))
print('Full validation gini: %.5f' % gini_normalizedc(ser_ytrain.values,
trprobs))
In [ ]:
# df_sub = pd.DataFrame({'id' : test_id, 'target' : txprobs})
# df_sub.to_csv('NN_EntityEmbed_10fold-sub.csv', index=False)
In [ ]: